And yet people of African ancestry are astonishingly underrepresented in the genetic reference panels used to inform the “ethnicity estimates” that DNA testing companies return to customers who send in spit samples, hoping to trace their origins to something specific enough to compare to documents and family lore. A wider representation in the reference panels would also aid the interpretation of genetic information in developing new precision medical treatments.
New hope for rectifying this imbalance, in the form of the sequencing of 426 African genomes, was announced at the recent 2019 American Society of Human Genetics annual meeting in Houston. The talk’s title encapsulated the vast scope of the work: “High-depth genome sequencing in diverse African populations reveals the impact of ancestral migration, cultural demography, and infectious disease on the human genome.”
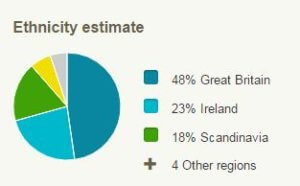 Right now, the colorful pie charts and polygons that depict customers’ geographic origins skew heavily European.
Right now, the colorful pie charts and polygons that depict customers’ geographic origins skew heavily European.
African origins
The trunk of the tree that led to humanity began in Africa about 6 million years ago. As time passed and some groups of people migrated out of Africa while others remained, mutations sculpted our modern genomes, yet always echoing our deep African roots. The greatest genomic diversity persists among Africans, which the new work confirms.
“There was a great deal of variation among people in the same region of Africa, and even among those from the same country. This reflects the deep history and rich genomic diversity across Africa, from which we can learn much about population history, environmental adaptation, and susceptibility to diseases,” said Neil Hanchard, MD, PhD, of the Baylor College of Medicine, when he presented the findings of the 426 sequenced African genomes at the meeting.
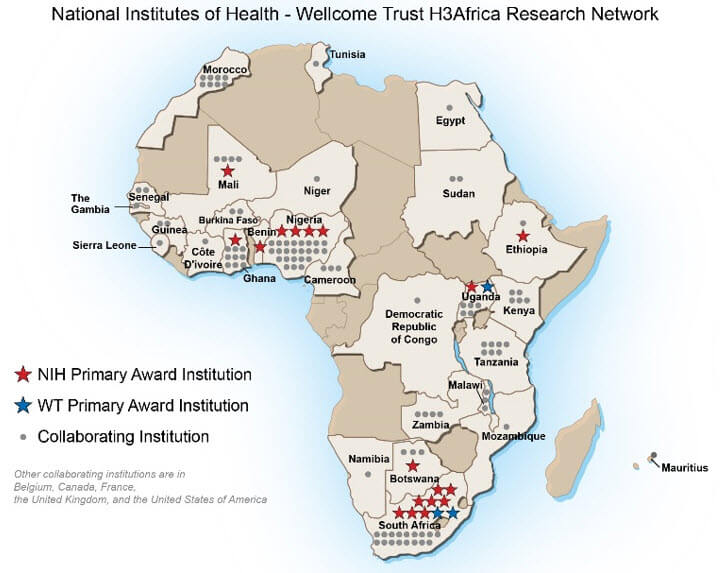
The research is part of the Human Heredity and Health in Africa (H3Africa) consortium, which began in 2012 as a partnership among the US National lnstitutes of Health (NIH), the African Society of Human Genetics, and the UK’s Wellcome Trust. African researchers are heavily involved in and lead the dozens of projects. “There is a dearth of baseline genetic data for African populations,” said Dr. Hanchard.
(I attended a meeting leading up to the genesis of H3Africa, in 2011 in Cape Town. After two days of trying to talk to one of the leaders, Charles Rotimi, director of the Center for Research on Genomics and Global Health at the NIH, I finally caught up to him – in front of Nelson Mandela’s jail cell on Robben Island, where we were allowed in two at a time. Of course we were speechless.)
 H3Africa’s goal is to use genomic strategies to dissect the underpinnings of disease and identify risk factors. Targets include rare single-gene diseases, more common conditions with complex genetic and environmental components like heart and kidney disease, and communicable diseases.
H3Africa’s goal is to use genomic strategies to dissect the underpinnings of disease and identify risk factors. Targets include rare single-gene diseases, more common conditions with complex genetic and environmental components like heart and kidney disease, and communicable diseases.
The consortium also supports investigations into the ethical, legal and social implications of genomics research; training; utilization of bioinformatics; biobanking; pharmacogenetics; and coordination and networking in conducting research and providing clinical services.
Africa: The “cradle of human genetic variation”
The researchers sequenced the genomes of 426 individuals from 50 ethnolinguistic groups from 13 African countries. Of those genomes, 323 were sequenced at high depth (many times), to increase the granularity of the findings and reveal rare gene variants. Then bioinformatics digested the data to deduce:
- patterns of admixture (how population groups intermingled over time)
- signs of natural selection (the persistence of helpful gene variants through reproduction and weeding out of harmful variants through infertility and early death.)
- distributions of “rare, novel, and medically important variation” in genomes
Overall the researchers found more than 3 million places in the genomes that vary in the DNA base among the African genomes. For example, at a certain point in a certain chromosome in a certain population, 90 percent of the people have the DNA base A (adenine), and the other 10 percent have G (guanine). Another population might have different proportions, or perhaps even different bases.
 These distinctions are called SNPs (for “single nucleotide polymorphism”; a nucleotide includes a DNA base and “polymorphism” means “many forms”). Comparing thousands or even millions of SNPs among individuals chosen for how they differ in a specific characteristic – a disease, a trait, or a country of origin, for example – forms the basis of many genetic tests and investigations, from identifying risk factors for disease to matching relatives.
These distinctions are called SNPs (for “single nucleotide polymorphism”; a nucleotide includes a DNA base and “polymorphism” means “many forms”). Comparing thousands or even millions of SNPs among individuals chosen for how they differ in a specific characteristic – a disease, a trait, or a country of origin, for example – forms the basis of many genetic tests and investigations, from identifying risk factors for disease to matching relatives.
(SNPs are also called SNVs, for single nucleotide variants.)
Mapping migration
Researchers impose broad time stamps on DNA sequence data using known rates of mutation for certain well-studied genes. Combined with knowing the homes of indigenous groups, these “molecular clocks” can shed light on ancient migration patterns. For example, newly identified SNPs from people from Mali revealed a paternal line from northern populations as well as telltale signs of inbreeding – places along the two chromosomes of a pair that are identical in DNA sequence.
Like adding new highways to a roadmap, the new genome sequences fill in some of the routes that people took out of East Africa, where fossils of our forebears go back 6 million years.
 “For the first time, our data from East and West Africa showed evidence of movement that took place 50 to 70 generations ago from East Africa to a region in central Nigeria. This movement is reflected in the genomes of a Nigerian ethnolinguistic group and is distinct from previous reports of gene flow between East and West Africa,” said Adebowale Adeyemo, MD, deputy director of the Center for Research on Genomics and Global Health at the National Human Genome Research Institute, and a senior author on the paper that will be published on the new study.
“For the first time, our data from East and West Africa showed evidence of movement that took place 50 to 70 generations ago from East Africa to a region in central Nigeria. This movement is reflected in the genomes of a Nigerian ethnolinguistic group and is distinct from previous reports of gene flow between East and West Africa,” said Adebowale Adeyemo, MD, deputy director of the Center for Research on Genomics and Global Health at the National Human Genome Research Institute, and a senior author on the paper that will be published on the new study.
The findings also added detail to the southern migration of the Bantu people, which was distinct from the whereabouts of the indigenous Khoisan, the modern people whose roots go back the farthest and are therefore the most genetically diverse. The Khoisan, also known as “San” or “Bushmen,” live in the Kalahari Desert in Botswana and Namibia.
The Khoisan and Bantu aren’t new to genomic scrutiny. An oft-cited paper from 2010 in Nature compared the complete genome sequence of a Khoisan man named !Gubi to that of Bantu South African civil rights activist Archbishop Desmond Tutu and to partial genome sequences of three other Khoisan who live near each other. The Khoisan genomes were as different from each other as a modern European genome is from that of a modern Asian genome. The four Khoisan genomes differ from the genome of Desmond Tutu at more than a million places.
The Khoisan gene variants reflect their hunter-gatherer lifestyle. A variant of the actinin-3 muscle gene promotes sprinting over distance running and a gene variant that encodes a chloride channel in cells conserves water, beneficial in the desert. The people use their “bitter taste” gene variant to detect poison and identify plants with medicinal value.

Gene variants that aren’t present provide information too. Khoisan genomes do not have the gene variant that protects other populations from malaria in carriers – but they don’t need it, because the mosquitoes that spread the disease can’t survive in the desert. Individuals with two copies of the gene variant have sickle cell disease and don’t survive to reproduce, removing it from the population. Other examples of genetic disease protecting against infection are here.
Disease clues
The new African genomes reveal 63 genes that emit “signals” of natural selection – specific DNA base sequences that alter the encoded proteins in ways that are advantageous and aren’t in the genomes of other primates. Thirty-three of the 63 genes affect the immune response to viral infection.
“When you consider which forces have shaped the genetic diversity of Africans, you tend to think of mosquito-transmitted diseases like malaria. Our study suggests that viral infections have also helped to shape genomic differences between people and groups, via genes that affect individuals’ disease susceptibility,” Dr. Hanchard said.
Single-gene mutations known to cause diseases in non-African populations appeared in less than 2 percent of the African genomes. The researchers looked for 59 conditions that the American College of Medical Genetics and Genomics recommends clinicians reveal to patients if they turn up “incidentally” on testing for another reason, because the conditions are treatable.

That’s good news. But each sequenced African genome harbored on average 7 “pathogenic” gene variants from another list, that of the NIH’s database ClinVar.
Some ClinVar variants were 10 times as prevalent among the African genomes as in the mostly European genomes upon which ClinVar is based. Their prevalence suggests that they’re not as dangerous against the backdrop of an African genome. Clues to how a gene variant is harmful in one population but not in another could suggest treatment strategies. (See Are Eurocentric Genetic Databases Hampering Health Care?)
The frequencies of gene variants behind diseases long-associated with African ancestry varied widely by geography. These include the distinctive mutation that causes sickle cell disease and the G1 and G2 variants of the apolipoprotein (APOL1) gene, responsible for a form of kidney disease. Like the sickle cell-malaria connection, the APOL1 variants persist in carriers because they destroy the parasites that cause African sleeping sickness.
Good news for consumer ancestry testing?
The researchers cited plans to use the new findings in precision medicine, to flesh out details of migrations and human history, and to reveal basic characteristics of population genetic structures. But the news release and abstract describing the project omit an immediately compelling application: in consumer DNA ancestry testing.
I hope the information from the hundreds of newly sequenced genomes will find its way into the databases of the ancestry testing companies, where African representation is ridiculously inadequate, especially considering the irony of humanity arising in Africa. Because the transatlantic slave trade obliterated knowledge of geographic origins for so many people, traditional genealogical sources, like ship manifests and docking points and sale and emancipation documents, are critically important. More genome information promises to fill in some gaps.
To see just how bare the African background is in consumer testing, I consulted the latest version of AncestryDNA’s reference panel. It’s derived from DNA samples from indigenous populations around the world – by definition, people who’ve lived there for as long as anyone or any records indicate. The reference panel lies behind the pretty pie charts and polygons depicting countries of origin.
The companies compare about 700,000 SNPs in customers’ samples to those from people in the indigenous populations in several iterations, each time omitting one reference population. If omitting a population doesn’t alter a customer’s results, then that population isn’t in the individual’s background. That’s why when companies add populations, people’s pie charts and polygons can shift.
The current reference panel represents 16,638 individuals, 1,395 of whom are indigenous Africans – that’s about 8.4%. As of 2017, Africans comprised 16.6% of the world population, a percentage expected to swell to 39.4% by 2100, due to a “youth bulge.”
That’s not terrible. But consider the flip side. Europeans account for 64.5% of the DNA samples that form the reference panel, but only 11% of the total world population.
I hope that the newly-sequenced African genomes, and more to come, will fill in the outlines of the origin and diversification of humanity.
Ricki Lewis is the GLP’s senior contributing writer focusing on gene therapy and gene editing. She has a PhD in genetics and is a genetic counselor, science writer and author of The Forever Fix: Gene Therapy and the Boy Who Saved It, the only popular book about gene therapy. BIO. Follow her at her website or Twitter @rickilewis