Cholera is an infectious disease of the small intestine caused by strains of the bacterium Vibrio cholerae. The classic symptoms of the disease include extensive bouts of acute diarrhea that can last several days and consequently lead to severe dehydration. This can lead to death if left untreated. Symptoms may start as little as two hours after ingestion of water contaminated with this bacterium. This short ‘incubation time’ is thought to be the reason for cholera being able to spread so rapidly through populations.
Researchers have estimated that there are up to 4 million cases of cholera every year and up to 140,000 deaths worldwide, so it is still very much a global health issue.
John Snow and the Broad Street water pump
The first cases of cholera in England were reported in 1831, around the same time an 18-year-old man called John Snow was completing his medical studies in London (he’d begun training as a medic at just 14 years old!). Over the next 20 years cholera caused a series of serious epidemics, killing tens of thousands of people in England alone.
Back then very little was known about how infectious diseases spread or even what infectious diseases were. At that time people believed that diseases like cholera and the Black Death were caused by breathing in miasma or ‘bad air’ coming from decomposing matter. However, the investigations of John Snow were about to challenge these ideas.
Although he specialised in women’s health and pregnancy, Snow was interested in many areas of medical science. He was particularly fascinated with how infectious diseases, like cholera, were spread. Since beginning his medical training, he was always keen to investigate water as a vehicle for transmitting infectious disease. In the mid-1800s, people didn’t have running water or clean ways to dispose of or treat sewage. London was particularly bad, with sewage often being dumped into open pits called ‘cesspools’ or even directly into the River Thames. In addition to this, water from the Thames was commonly bottled and delivered to pubs and other businesses for consumption!
Snow recognized this and suspected that sewage could be contaminating the water supply and spreading cholera, and probably many other diseases, around the city.
In September 1854 a particularly severe outbreak of cholera hit the Soho area of London, close to where he lived. He took the opportunity to find the source of the outbreak, once and for all. He worked around the clock to track the infection by examining hospital and public records.
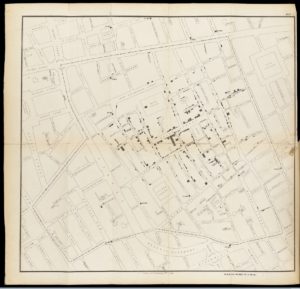
Snow constructed a map (below) showing the location of various water pumps around the city and where deaths from cholera were clustered. He showed the number of deaths at each address as a series of horizontal lines, stacked up like a pile of bodies in the street.
Some groups of people in the area had managed to avoid cholera despite living and working near the pump so he investigated why this was. He found one group of men working in a brewery on Broad Street who had remained healthy by avoiding drinking water from the pump and instead sticking to their own beer. Although unbeknown to them at the time, the fermentation process kills the cholera bacteria so drinking beer and gin was actually much safer than drinking water back then!

John Snow’s cholera map is an important early example of data visualisation and represents a significant milestone in the birth of the field of epidemiology – the branch of medicine that investigates the incidence, distribution and control of diseases.
On 7th September 1854 Snow took his findings to the town officials and convinced them to take the handle off the Broad Street water pump. Although they initially refused his request, after removing the handle the outbreak of cholera almost immediately dissipated.
A few decades later, the German physician Robert Koch identified the cause of cholera, the bacterium Vibrio cholerae. Koch confirmed that the bacterium was indeed spread via unclean water or food, providing concrete support for John Snow’s theory. Snow is now widely credited with establishing the field of epidemiology. To mark the importance of his discovery, the Broad Street pump is on permanent display in the London School of Hygiene and Tropical Medicine.
Still a disease in the modern era
Many people think that the cholera story ends there, but it is a still very much a major public health issue. Today, cholera causes millions of cases of diarrhea and hundreds of thousands of deaths every year. Even in the UK there are about 12 cases of cholera reported every year, although these are generally associated with overseas travel to developing countries. Since the 1880s, developed countries like the UK have built up a solid infrastructure for the distribution of clean water, as well as the disposal and treatment of sewage. This has prevented the transmission of water borne diseases such as cholera. But there are still many resource-poor areas of the world where sanitation remains a major problem.
Cholera is still relatively common in areas of the world where there are no clean water and sewage disposal systems such as in parts of Africa, Asia and South America. However, outbreaks of cholera can still occur in other parts of the world. For example, there was an outbreak of cholera in Haiti in 2010. This was after a devastating earthquake that struck the island of Hispaniola, which is made up of the Dominican Republic in the east and Haiti in the west. The earthquake killed more than 160,000 people and caused a loss of infrastructure to Haiti that led to political and social unrest. This loss of infrastructure contributed to a devastating cholera outbreak.

Tracking cholera through genomics
In the past, scientists have used fairly blunt tools to investigate cholera, but with DNA sequencing? we can now gain a much deeper understanding of the bacterium by looking at its genome?.
There are many different types of cholera, but only a few that cause “pandemic” disease. This refers to the types of cholera that have spread rapidly across countries and continents. Six cholera pandemics have been recorded since 1816. The current, seventh pandemic is caused by a type of cholera bacteria named after the village where it was originally found, El Tor in Egypt. Pandemic types of cholera are of primary interest for DNA sequencing because they cause the most widespread and devastating disease.
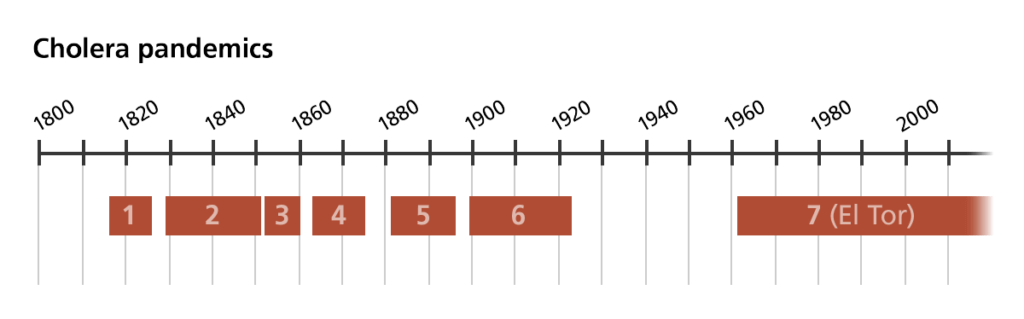
To track the spread of cholera across the globe, scientists can sequence the genomes of thousands of samples of cholera bacteria taken from different people from different regions of the world. These are called ‘isolates’. By comparing the genomes of these isolates, scientists can now say with confidence that different cholera bacteria from different countries are related to each other. They can also work out how closely related different isolates are and how recently they separated from each other. They can tell whether they separated from each other just a few days or weeks ago or whether they are more distantly related and separated by a few years or even decades.
This is useful information to know. If two people are found to be infected with cholera bacteria that are closely related, then it may suggest that they have both recently visited the same country or community and may even have drunk from the same contaminated water source. If that’s the case then it is clear where the origin of the cholera infection is and attention can be focused on eliminating that source of the disease.
Identifying outbreak origins
Sequencing a genome enables us to find out the exact order of DNA bases in a genome. If we sequence lots of genomes then we can then compare them side by side to find the similarities and differences. This is what scientists have done with cholera. Primarily they have been looking for single base differences in the genomes from different isolates. The fewer single base differences there are between two genomes, the more closely related those isolates are to each other.
By comparing lots of genomes in this way, scientists can draw a phylogenetic tree. Like a family tree, a phylogenetic tree shows how different individuals are related to each other. For example, comparing the genomes from isolates of cholera enables scientists to establish a tree of how these isolates are related to each other.
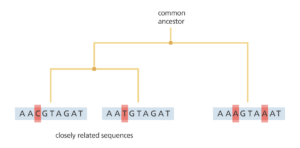
Scientists analysed the DNA from cholera isolates from 1930 up to the present day. From this they created a phylogenetic tree for cholera showing the relationships between the isolates causing the current pandemic. By analyzing the tree, they identified a single, evolving population of Vibrio cholerae that acts as the source of all cholera. All cholera outbreaks around the world can be traced directly back to this source population, but each local outbreak eventually dies out and isn’t seen again. However whilst these individual outbreaks die out, the source population persists and continues to evolve. So is this source population found in one place or is it moving around?
To answer this question, scientists at the Wellcome Trust Sanger Institute looked at where each isolate came from. What they saw was that the cases of cholera from South Asia all branch off directly from the main source population. In contrast, the outbreaks in other parts of the world like Africa and South America were linked to this source population but were slightly more distantly related to it. This suggests that the South Asian isolates are more closely related to the source population, pointing to the location of the source also being in South Asia. Further investigation identified this source location as being in the Bay of Bengal, positioned in the middle of India and Sri Lanka, Bangladesh and Myanmar (Burma). This stretch of water may well be the world’s equivalent of the Broad Street Pump identified by John Snow, from which the world’s cholera pandemics stem.
What next?
With this evidence scientists now know they need to focus their efforts on tackling cholera in the Bay of Bengal and essentially, take the handle off the theoretical water pump. By doing this they can hopefully prevent any further spread of cholera to the rest of the world and grasp a much stronger understanding of the current cholera pandemic.
However, this will require resources, funding, and lots of cholera samples. It will also take time to fully understand how the cholera is spreading and the nature of the water sources that are involved. This will ensure that any vaccination or drug campaign is carried out effectively, efficiently and gives cholera the knock-out punch.
Building on the foundation of John Snow’s work all those decades ago, we now have an extremely powerful tool using genomics to make high resolution maps to show how a whole host of infectious diseases spread. These maps can be used to understand outbreaks across regions, countries and continents but also in specific local communities and hospitals. Hopefully they will enable scientists and doctors to gain the upper hand when tackling infectious diseases like cholera in the future.
A version of this article was originally published on Your Genome’s website as “Science in the time of cholera” and has been republished here with permission.
































