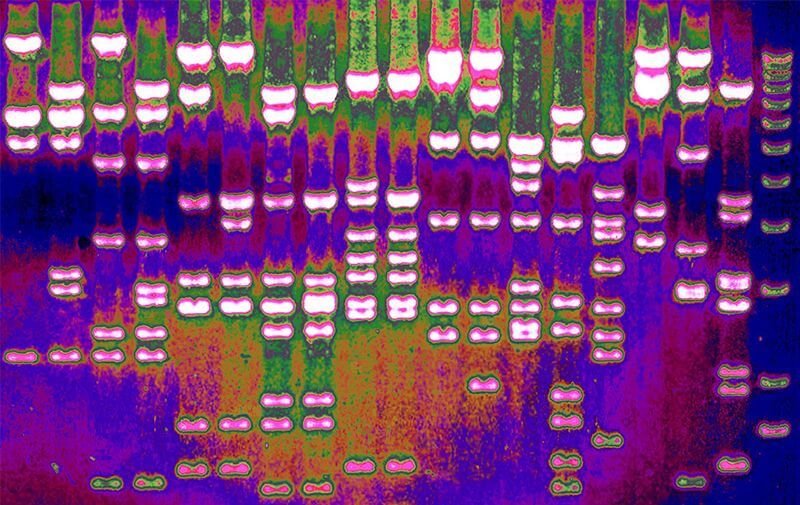
Each person’s genome contains three billion building blocks called nucleotides, and researchers must compile data from thousands of people to discover patterns that signal how genes have been shaped by evolutionary pressures. To find these patterns, a growing number of geneticists are turning to a form of machine learning called deep learning.
…
One deep-learning tool called ‘DeepSweep’, developed by researchers at the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, has flagged 20,000 single nucleotides for further study. Some or all of these simple mutations may have helped humans survive disease, drought or what Charles Darwin called the “conditions of life.”
…
One example is the mutation that gives many adults the ability to drink cow’s milk. It enables the body to produce lactase, an enzyme that digests the sugar in milk, into adulthood. By analysing human genomes with statistical methods, researchers discovered that the mutation spread rapidly through communities in Europe thousands of years ago — presumably because nutrients in cow’s milk helped people to produce healthy children.
…
DeepSweep’s creators trained their algorithm on signatures of natural selection that they inserted into simulated genomes. When they tested it on real human-genome data, the algorithm zeroed in on the lactase mutations that allow adults to drink milk. That bolstered the team’s confidence in the tool.
Read full, original post: Machine learning spots natural selection at work in human genome