A Chinese biologist raised the hopes of the scientific community last year when he announced the discovery of a gene editing tool with the potential to upstage the transformational CRISPR-Cas9 system. Han Chunyu, of Hebei University of Science and Technology, dubbed his technique NgAgo — for Natronobacterium gregoryi Argonaute.
But his promise of a more accurate gene editing tool has been followed by a steady stream of disappointments, with other scientists failing to reproduce Han’s results. Mounting criticism has prompted Nature Biotechnology, the research publisher, to investigate the matter.
The work of Han’s research team is significant on multiple fronts. For China, it represented a first, in terms of that nation’s contribution to the realm of gene editing. But on a worldwide scale, it offered the potential of a precise gene editing tool with a wide range of applications, and fewer off-target effects.
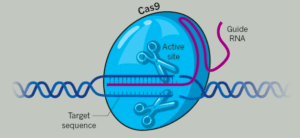 The popular CRISPR system has been likened to a pair of molecular scissors that uses the enzyme Cas9 to essentially cut or insert small pieces of DNA. Scientists have used CRISPR to edit genes in a multitude of applications, including adding and removing hypertension in rats to providing a potential solution to citrus disease.
The popular CRISPR system has been likened to a pair of molecular scissors that uses the enzyme Cas9 to essentially cut or insert small pieces of DNA. Scientists have used CRISPR to edit genes in a multitude of applications, including adding and removing hypertension in rats to providing a potential solution to citrus disease.
Han described NgAgo to China Central Television using a similar analogy:
[CRISPR] is a pair of golden scissors, and people are thinking how to make it even shinier. But there are other kinds of scissors, and what we found is a new one. Whether it is good or not, it is still a new pair.
One main difference between the two pairs of “scissors” is that the NgAgo system uses an Argonaute endonuclease instead of the Cas9 enzyme to guide the cutting. As a result, it is less likely to be confused by free floating single-stranded DNA which has been known to cause inaccuracies in the CRISPR edits.
When NgAgo was first announced, many scientists celebrated the discovery, touted by Hebei University of Science and Technology for its novelty.
The university told the Global Times:
NgAgo is the first Chinese-invented top-notch biotechnology, breaking the foreign monopoly on genome editing processes. The technology has extensive applications for agriculture and medicine, including in gene therapy for diseases like AIDS and hepatitis B.
Some researchers suggested that NgAgo had the potential to be much more accurate than CRISPR-Cas9.
George Church, a geneticist at Harvard Medical School in Boston, told Nature: “A lot of us are really cheerleading and hoping that it works.”
To date, only a few scientists have been able to successfully use the technique since it was first published in Nature Biotechnology on May 2, 2016.
Controversy first went international in July, shortly after Australian geneticist, Gaetan Burgio, posted a detailed attempt of his failure to replicate Han’s NgAgo experiment on his blog.
However, Nature reported that at least three scientists, all of whom wished to remain anonymous, have had some success with the technology.

One Chinese researcher, independent of Han’s team, tested NgAgo in a few kinds of cells and found that it was able to induce genetic mutations at the desired sites. The researcher has verified the findings by sequencing. Two more Chinese scientists say they have initial results showing that NgAgo works. However, they still need to confirm this with sequencing.
Yet even if these few successes are proven true, Burgio argues that the difficulties encountered by other researchers suggest the technology may never fulfill its initial promises. He told Nature: “It might, might work, but if so, it’s so challenging that it’s not worth pursuing. It won’t surpass CRISPR, not by a long shot.”
Criticism for NgAgo has been growing steadily since Burgio’s blog post as more and more scientists have reported failures at reproducing the original results, according to Retraction Watch. Much of the backlash was directed at Han for not adequately addressing the issues scientists have been experiencing with reproducibility.
Han initially suggested that other scientists might be having trouble duplicating his results because they required “superb experimental skills.” He also said the failures could be related to the contamination of test cells by a bacteria called Mycoplasma.
Han’s defenses have been dismissed by critics. Researchers from eight US and Chinese laboratories published a letter, on November 15, in the online issue of Protein & Cell, which stated:
The key point of [Han and his colleagues paper] is that DNA-guided NgAgo’s can efficiently target 47 genomic loci with a 100 [percent] success rate and [greater than] 20 [percent] efficiency. Neither the originally published protocol nor the newly released information on Addgene’s website involves any steps that seem to require “superb experimental skills.”
…
…it [also] seems unlikely that independent laboratories would all have their cells contaminated, resulting in consistently negative results for DNA editing activity. In fact, several of the signees of this letter have made sure that our cells are free of mycoplasma by first testing them before performing replication experiments.
Another letter was published on November 28 in Nature Biotechnology that offered similar criticism to Han’s claims that NgAgo can be used as a gene editing tool.
These issues have left the scientific community clamoring for Han and his team to clear up the uncertainty surrounding NgAgo. The criticism has prompted Hebei University to launch an inquiry. Nature Biotechnology, the journal that originally published the NgAgo technique this past May, has also begun investigating.
On November 28, the journal issued an editorial expression of concern:
Nature Biotechnology believes that it is important for authors to be able to investigate the concerns raised by the correspondence and to provide additional information and evidence to support their paper if they are able to do so. Thus, we will continue to liaise with the authors of the original paper to provide them with the opportunity to do that by January 2017. An update will be provided to the community at that time.
Elizabeth Newbern is a contributing writer for the GLP. She has also written for GenomeWeb, Live Science, Audubon Magazine, and Scholastic. She received her MA in Journalism from New York University and a BA in Geology from Bryn Mawr College. You can follow her on twitter @liznewbern.































