Imagine you could scan a child’s face with a phone app, as quickly as Star Trek’s Dr. McCoy could wave his medical tricorder around a patient, and get an instant diagnosis of a rare genetic disease. You can’t actually do it yet, but facial recognition technology is rapidly advancing. Police use it to spot suspected criminals, government agencies use it to find and track terrorists, and casinos use it to compare faces of people who might be cheating with faces in a cheating database.
The technology also is working its way into clinical medicine, particularly in the area of medical genetics. There are some glitches to be solved, to be sure, notably when it comes to recognizing facial peculiarities linked to syndromes based mostly on genetic data from populations of European decent. Technological solutions are now showing up for this issue, but sometimes portrayal in the media can be confusing. Such was the case following publication of a study showing that facial recognition can help physicians identify a rare disorder called DiGeorge syndrome.
Multiple takes on one story
Stories about computer recognition of DiGeorge syndrome facial features offered different takes on the subject. Some suggested the program could diagnose the condition, while others were more subdued, saying the program could simply aid in diagnostics. Most of the stories, spawned by a study published in the American Journal of Medical Genetics, conveyed an important point — that facial-scanning technology can allow detection of features of some genetic diseases in diverse population groups, as opposed to just in homogeneous populations, and that this capability can help physicians.
But some of the stories lost sight of the fact that the study showed only how the technology works for a particular rare disease, 22q11.2 deletion syndrome, also called DiGerorge syndrome. Resulting from the absence of a section of chromosome 22, DiGeorge syndrome is inherited in a dominant pattern (a child gets it from either one or both parents) and manifests notoriously with problems in the heart, immunity, low blood calcium and low parathyroid hormone levels, and a distinguished facial appearance. That last feature is why DiGeorge syndrome is one of a few genetic disorders that match up with databases of facial features on a couple of different algorithms. One algorithm was developed by the National Institutes of Health and the other, called Face2Gene, by a company called FDNA. The result is that facial recognition technology can identify a person with DiGeorge syndrome 96.6 percent of the time. The study also demonstrated that the technology can identify the condition with much better accuracy in non-Caucasians than earlier incarnations of the technology could. This is key, because there’s a paucity of medical genetics specialists in developing countries, so the system might be useful in telemedicine applications.
But if you look at the most popular general interest articles published as a result of the paper, the message is muddled, leaving readers with the impression that the technology is ready for a wider swatch of disorders.
Variations
The original press release, put out by the National Human Genome Research Institute (part of the National Institutes of Health, opened with this perspective:
Researchers with the National Human Genome Research Institute (NHGRI), part of the National Institutes of Health, and their collaborators, have successfully used facial recognition software to diagnose a rare, genetic disease in Africans, Asians and Latin Americans. The disease, 22q11.2 deletion syndrome, also known as DiGeorge syndrome and velocardiofacial syndrome, affects from 1 in 3,000 to 1 in 6,000 children.
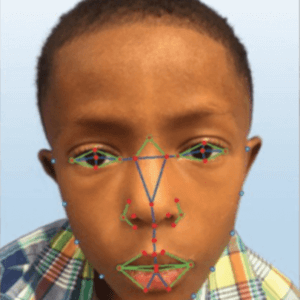 The press release went on to note that the novel finding of the study is that the technology can work well to identify facial features of DiGeorge syndrome in non-Caucasian populations, and that the app learns by experience. Thousands of images are fed into the system, so it learns from experience — just like a medical geneticist learns by seeing face after face. The press release also noted the same research team previously demonstrated almost as good accuracy for the system in identifying facial features associated with another genetic disorder, Down syndrome, but that studies had yet to be done on its accuracy for detecting facial features of two other conditions: Williams syndrome and Noonan syndrome.
The press release went on to note that the novel finding of the study is that the technology can work well to identify facial features of DiGeorge syndrome in non-Caucasian populations, and that the app learns by experience. Thousands of images are fed into the system, so it learns from experience — just like a medical geneticist learns by seeing face after face. The press release also noted the same research team previously demonstrated almost as good accuracy for the system in identifying facial features associated with another genetic disorder, Down syndrome, but that studies had yet to be done on its accuracy for detecting facial features of two other conditions: Williams syndrome and Noonan syndrome.
As days passed, the story started morphing as different publications juggled sections of the press release and added their own explanations. In Real Clear Life, for example, the juggling produced this section:
To fix this, researchers at the NHGRI developed a method that uses facial recognition to detect DiGeorge condition within 96.6 percent accuracy. The technology uses a combination of machine learning and computer vision, a type of image analysis, to generate 126 characteristics the software looks for. Similar technology has already been used to diagnose Down syndrome.
This implies that the system provides diagnoses, but actually the system being tested in the new study, Face2Gene, only provides probabilities which physicians can then weigh against their experience to make their own diagnosis. This may sound like a minor point, but in the original press release, Down syndrome was mentioned as an example of a condition for which accuracy already had been worked out in terms of probabilities.
“We are not a diagnostic tool, and we will never be a diagnostic tool,” said FDNA CEO Dekel Gelbman.
Genetic disease application
Facial recognition capability is advancing on all fronts, but it’s noteworthy that the challenges for medical genetics actually are simpler in comparison with criminal justice applications. An analysis published on physorg points out the following that would come into play for security applications:
Not only would such a system require millions of cameras capable of producing high-quality footage, but it would also require the integration of photo-ID databases such as mugshots from every police force, previous passport images, and driving license images for everyone in the country.
In contrast, Face2Gene and another app employed by NIH researchers match facial features against averages of measurements known to be associated with all individuals sharing a particular genetic condition. This makes a match more likely compared with a match between features being used to connect a single individual with potentially millions of candidates. The NIH system actually gives out diagnoses, while the Face2Gene supplies just probabilities. But in both cases, the capability is there for clinicians to use routinely, at least when it comes to DiGeorge and Down syndromes. Very soon similar studies will look at the same approach to Noonan syndrome and Williams syndrome, both of which are genetic conditions where facial features play significantly into the diagnosis.
David Warmflash is an astrobiologist, physician and science writer. BIO. Follow him on Twitter @CosmicEvolution.