The SEC thumb’s up may be a first, but it follows in the footsteps of Nebula Genomics, which Harvard’s George Church founded in 2016. The Nebula strategy: “Get sequenced. Discover your DNA and ancestry. Become one of the first to profit from your own genomic data.” (However, the public launch of LunaDNA was in December 2017, but Nebula in February 2018).
Both celestially-named companies connect consumers who have access to their DNA findings to researchers whose projects require a critical mass of data. Everyone wins. And in the process, the economics of human DNA sequencing has completely turned around. DNA data are now information for sale.
Remember the decade when sequencing the first human genome came with a $2.7 billion price tag?
Two technological leaps catalyzed the reversal in the valuation of DNA data: vastly accelerated genome sequencing and the rise of consumer genetics. Both happened in the 2007-2008 time period.
From Big Science project to personal asset
Much has changed since sequencing a human genome began to transition from science fiction to science reality in the mid 1980s.

I attended one of the first meetings to discuss sequencing “the” human genome, in Boston, in 1987. The initial cost estimate: about $1 billion, given the technology of the time. The National Human Genome Research Institute (NHGRI) graphically tracks the cost (see graphic).
Illumina, founded in 1998, is largely behind the sharp drop in sequencing costs that began in 2007. Their next-generation sequencing replaced Celera Genomics’ “shotgun” approach, which had replaced the NHGRI’s slow chromosome-by-chromosome assembly technique, with microfluidics devices that enabled massively parallel sequencing. It’s a little like comparing reading a book’s chapters, slowly, one a night (NHGRI) to meticulously reading all the way through (Celera) to capturing all the pages at once and your brain rapidly knitting them together (Illumina).
A drop in price was inevitable, reminiscent of the first calculators selling for $100 in the 1970s dropping to a few dollars, and now a free phone app. Illumina foresaw, back in 2003, the day when sequencing a human genome would cost $100.
Thanks to next-gen sequencing as well as major funding to improve technology, the cost of human genome sequencing blasted past the predictions of Moore’s law in 2008. Gordon Moore, the co-founder of Fairchild Semiconductor and Intel, in a paper in 1965 described the doubling each year of the number of components in an integrated circuit, which he revised to a doubling every two years. The law was later applied to computing power and then fit the declining cost of genome sequencing – until 2007.
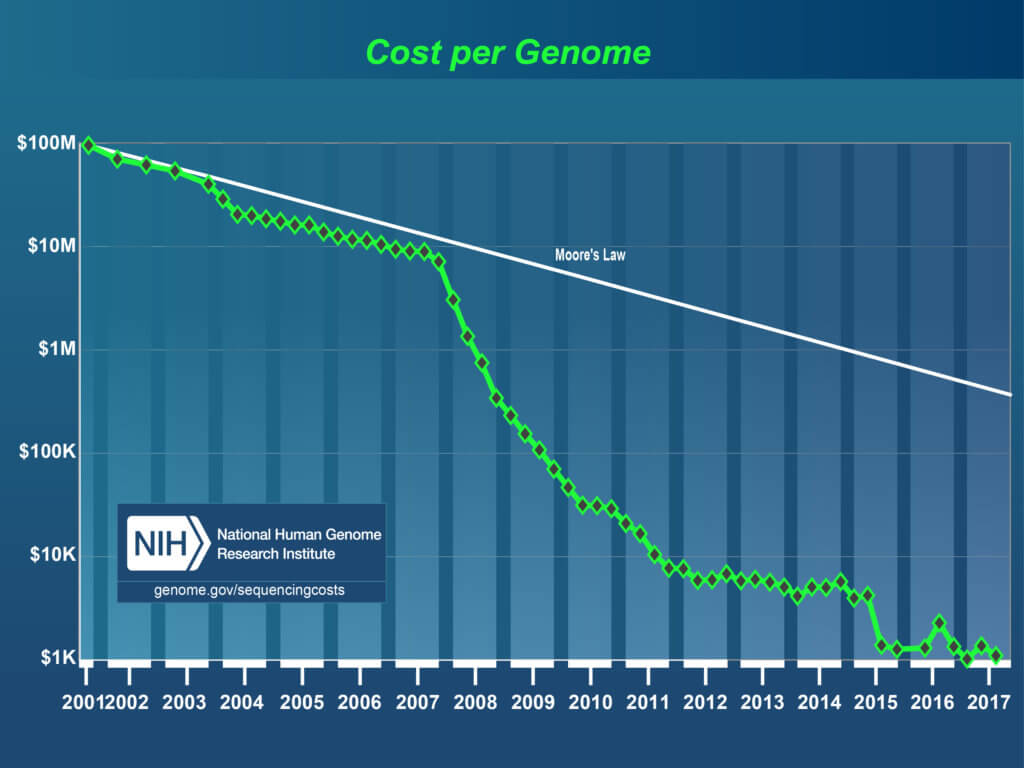 From then on, many a talk at a genomics meeting has begun with the iconic plunging slope of sequencing costs below the predictive curve of Moore’s law. “The price of sequencing an average human genome has plummeted from about US $10 million to a few thousand dollars in just six years. That does not just outpace Moore’s law — it makes the once-powerful predictor of unbridled progress look downright sedate,” wrote Erika Check Hayden in Nature.
From then on, many a talk at a genomics meeting has begun with the iconic plunging slope of sequencing costs below the predictive curve of Moore’s law. “The price of sequencing an average human genome has plummeted from about US $10 million to a few thousand dollars in just six years. That does not just outpace Moore’s law — it makes the once-powerful predictor of unbridled progress look downright sedate,” wrote Erika Check Hayden in Nature.
At the same time that human genome sequencing was becoming economically feasible, the companies offering genetic tests to the public emerged. It was alarming at the time. Here’s how I described the official announcement:
On a Thursday night in October 2007, I sat with hundreds of geneticists at the American Society of Human Genetics annual meeting in San Diego, so stunned that we ignored the free dessert. At a table in front of the crowd were several very nicely-dressed physicians and genetic counselors representing a trio of companies gearing up to offer, in the coming year, direct-to-consumer genetic testing.
Yes, ordinary people would be able to send samples of themselves – spit, it would turn out – to companies that would charge fees to return results right to them, circumventing health care professionals. The companies had names much catchier than those of the biotech companies of the past: 23andMe, Navigenics, (absorbed into Life Technologies Corp), and deCODE Genetics (now a subsidiary of Amgen). The chatter in the hallways afterwards, according to my unscientific survey, was negative.
No more.
Privacy concerns mount
 Kelly Ripa hawking AncestryDNA kits everywhere and the depleted pile of 23andMe boxes at my local Target indicate better than any study that consumer genetics is here to stay. But with all of that data flowing in — some of it exposing long-held family secrets or altering personal narratives — privacy has become an issue.
Kelly Ripa hawking AncestryDNA kits everywhere and the depleted pile of 23andMe boxes at my local Target indicate better than any study that consumer genetics is here to stay. But with all of that data flowing in — some of it exposing long-held family secrets or altering personal narratives — privacy has become an issue.
So I took a look at five places where people might deposit their DNA data.
23and Me (2008) Phrases like “de-identification/pseudonymization, encryption, and data segmentation” make privacy protection here seem airtight. Customers can delete their accounts and data at any time, and can choose which health reports to view, with whom to share information, and whether or not to consent for use of their information in the company’s research.
The 23andMe database quickly grew large enough to offer great statistical power. The company’s 2011 discovery of two genes associated with Parkinson’s disease was accomplished in record time using 3,426 cases and 29,624 controls. Their model of citizen science, once shocking, is clearly working.
GEDmatch (2010) This website burst into public consciousness in April 2018, with the use of its data to capture the Golden State Killer. But Curtis Rogers, a retired businessman and John Olson, a transportation engineer, conceived GEDmatch in 2010 as a tool for genealogists, professional and amateur. Participants upload their DNA ancestry data from consumer genetics testing companies (typically 770,000 or so markers), agree to a disclaimer, and receive a long list of their closest relatives who’ve uploaded their data.
I did it. Of my first 25 GEDmatches 16 have names, but some could be pseudonyms. Participants post their email addresses if they want to hear from half-sibs and missing cousins and such.
The privacy policy states that the purpose of the site is to share information, with a caveat added following the Golden State Killer capture: GEDmatch will provide data in compliance with a subpoena or warrant, and notify the owner of the DNA should this happen, if possible.
 All of Us (2015) The NHGRI’s Precision Medicine Initiative Cohort Program began after President Obama briefly mentioned personalized health care in his 2015 State of the Union address. A follow-up meeting at the White House in March 2015 led to development of a privacy statement. Yet again I was fortunate enough to attend an early meeting that introduced the program: an initial one million volunteers would provide health data for tracking and analysis, including blood samples so presumably DNA data.
All of Us (2015) The NHGRI’s Precision Medicine Initiative Cohort Program began after President Obama briefly mentioned personalized health care in his 2015 State of the Union address. A follow-up meeting at the White House in March 2015 led to development of a privacy statement. Yet again I was fortunate enough to attend an early meeting that introduced the program: an initial one million volunteers would provide health data for tracking and analysis, including blood samples so presumably DNA data.
By mid-2017, the Precision Medicine Initiative Cohort Program either spun off or renamed – I’m not sure which – the citizen-participation part as “All of Us.” It was as if somehow between 2015 and 2017 the American public suddenly couldn’t handle multisyllabic words or grasp intriguing concepts like precision medicine.
All of Us entails neither fees to participate nor compensation for doing so. Even Steven.
Nebula Genomics (2016) This company, already mentioned, is based on “blockchain technology,” invented in 1991 to apply cryptographic links with unalterable timestamps to a growing list of records (termed blocks). For Nebula that means connecting people holding information – consumers – to those who need it – researchers. “We shift the sequencing costs to researchers and enable people to get compensated for data sharing,” the company website states.
The “how it works” part at Nebula is, well, a little nebulous. A user “earns credits” by taking a survey, and can apply them “for valuable rewards.” Then the participant may be matched to a research project that pays for whole genome sequencing.
 But genome sequencing is also on sale at Nebula, this holiday season of course, for $99, which buys “0.4x coverage.” If I’m interpreting that correctly, it means 40% of the genome, a little like receiving four chapters of a ten-chapter book from Amazon – they might be the most informative chapters, but still, not a complete picture. High-quality coverage sequences the same genome several dozen times, to assure that all bases are counted. (A graphic on the site also reads “Gemone.”)
But genome sequencing is also on sale at Nebula, this holiday season of course, for $99, which buys “0.4x coverage.” If I’m interpreting that correctly, it means 40% of the genome, a little like receiving four chapters of a ten-chapter book from Amazon – they might be the most informative chapters, but still, not a complete picture. High-quality coverage sequences the same genome several dozen times, to assure that all bases are counted. (A graphic on the site also reads “Gemone.”)
Whatever the coverage, the results enable a participant to “understand your genome, explore your ancestry, and learn about your inherited traits,” rather vaguely stated as “appearance, personality, athleticism, nutrition, and sleep.” Like at 23andMe, disease risks, carrier status, and adverse medication responses are also part of the deal. One can then “continue to receive insights, analytics, and rewards on the platform” in exchange for consenting to use of the data.
LunaDNA (2017) The name honors the moon as a beacon that harkens back to the roots of company executives at Illumina. Rather than providing testing services, Luna works with health information that individuals already own, to accelerate research. A user uploads files from 23andMe, AncestryDNA, MyHeritage, FamilyTreeDNA, or others. In 2019 LunaDNA will also accept whole exome and genome files, RNA data, microbiome profiles, and info from electronic health records, fitness and activity trackers, and smart devices.
In an interview with the GLP, president and co-founder Dawn Barry explains the idea:
What if we invited individuals in, in a way that honors their needs for control of privacy and sharing in a responsible way? We get permission to use their longitudinal real world data and they share in the value of discovery.
Luna isn’t a marketplace to sell data, she says, but “a platform for organizing data for the greater good of health research. The data are deidentified and aggregated so research is done at the population level. In that process, identifiers are stripped away. You become a hashtag in the system. However, you have the ability to turn on a preference setting that you would like to be contacted for research.”
 For example, a researcher may seek women over 40 who have had breast cancer to recruit for a clinical trial. “If you’ve turned on preferences and fit the profile, we send out notification that someone is interested in working with you in a research capacity. Only you have control to break the anonymity of the system,” Barry adds. Matches will go far beyond cancer.
For example, a researcher may seek women over 40 who have had breast cancer to recruit for a clinical trial. “If you’ve turned on preferences and fit the profile, we send out notification that someone is interested in working with you in a research capacity. Only you have control to break the anonymity of the system,” Barry adds. Matches will go far beyond cancer.
Barry compares LunaDNA, which she sees as a health data co-op, to the other projects and companies. Nebula, she says, seems “more transactional” in that consumers complete surveys in exchange for low-cost low-coverage sequencing. And “the government effort doesn’t have a sharing element in the value created. You can’t control what type of research you want to be involved in.”
I’ll bet that the people at the 1987 meeting on sequencing the human genome and those on the stage at the genetics meeting two decades later introducing the first consumer genetic tests couldn’t have imagined the rise of citizen scientists supplying the data stream to drive precision medicine. Concludes Barry, “We celebrate all of the programs putting people at the center of research.”
Ricki Lewis is the GLP’s senior contributing writer focusing on gene therapy and gene editing. She has a PhD in genetics and is a genetic counselor, science writer and author of The Forever Fix: Gene Therapy and the Boy Who Saved It, the only popular book about gene therapy. BIO. Follow her at her website or Twitter @rickilewis