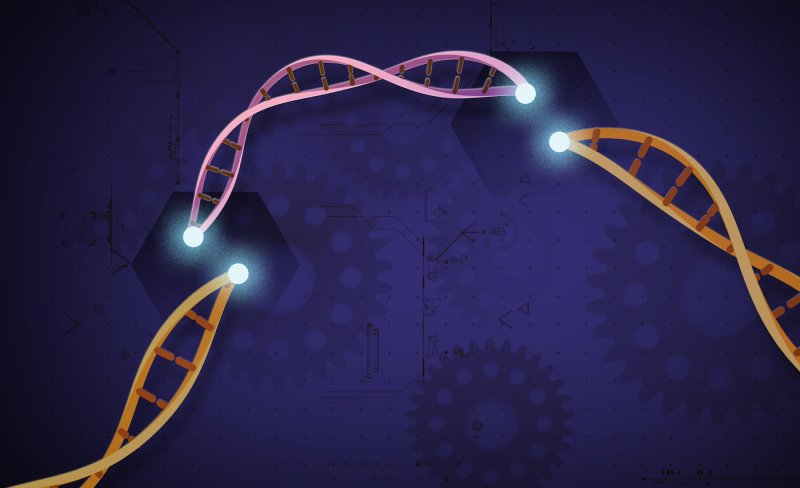
In search of new ways to sequence human genomes and read critical alterations in DNA, researchers have successfully used the gene cutting tool CRISPR to make cuts in DNA around lengthy tumour genes.
The researchers Johns Hopkins Medicine say this can be used to collect sequence information, and that pairing CRISPR with DNA sequencing tools, that sequence the DNA components of human cancer tissue, is a technique that could, one day, enable fast, relatively cheap, sequencing of patients’ tumours.
This could lead to streamlining the selection and use of cancer treatments that target highly specific and personal genetic alterations.
…
Winston Timp, PhD, assistant professor of biomedical engineering and molecular biology and genetics at the Johns Hopkins University School of Medicine, said: “For tumour sequencing in cancer patients, you don’t necessarily need to sequence the whole cancer genome. Deep sequencing of particular areas of genetic interest can be very informative.”
…
In addition to its potential to guide treatment for patients, Timp says the combination of CRISPR technology and nanopore sequencing provides such depth that it may help scientists find new disease-linked gene alterations specific to one allele (inherited from one parent) and not another.